The University of Gothenburg’s Dr Björn Burmann explains how his team’s research into bacteria is providing new insights into the functionality of DNA-repair proteins.
One of the fundamental cellular processes which ensures the living of all organisms, from simple bacteria to plants and mammalians, is the replication of our genetic material, the DNA. In the DNA of an organism, all the information is essentially ‘hard-coded’, thereby ensuring that all replicated copies are the same as the original. Of course, such a fundamental approach has to be heavily controlled to make sure that only correct copies are made. In addition to controlling the crucial feature of regulation directly at the replication level, our genetic material is also exposed to a myriad of environmental stresses as well, which can lead to the genetic material being damaged. In order to cope with this damage, cells have developed evolutionary conserved DNA-repair mechanisms. The importance of this is underlined by the fact that a malfunction in these essential repair machineries leads to cell death in bacteria, whereas in humans it can be the cause of skin cancer. This clearly highlights the fundamental importance of understanding these basic cellular processes at the molecular level and, indeed, of an even more detailed understanding at the atomic level.
Nucleotide excision repair
Evolution has come up with a variety of different solutions to screen DNA for errors, but for my team the coupling between different cellular machineries has been a long-standing interest (Burmann et al. 2010). Cells also employ the RNA-Polymerase to drive the transcription process, which transcribes the genomic DNA to RNA which then can be translated into proteins, as a sensor of DNA breaks, or adducts created by UV-light and metabolic agents (Truglio et al. 2006). Subsequently, the RNA-Polymerase recruits different DNA-repair proteins that cut out a short stretch around the DNA damage and replace it. The repair process is referred to as ‘nucleotide excision repair’ (NER). In bacteria, the NER depends on the interplay of the DNA-repair machinery, the so-called ‘UvrABCD’ proteins, recognising and cleaving damaged DNA in a multistep reaction under ATP consumption, a process which has been known for some time (Sancar and Rupp, 1983).
Mfd
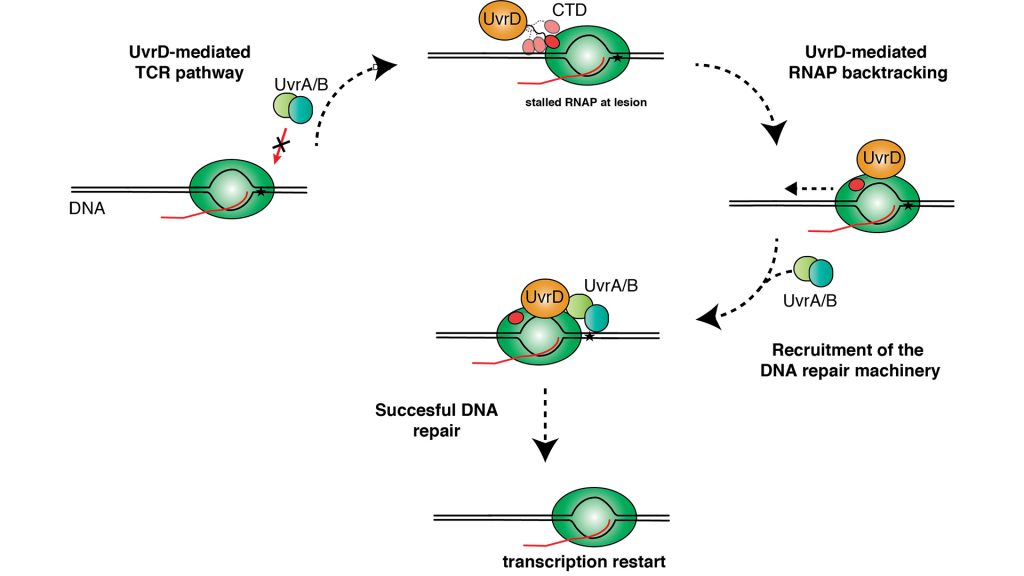
By using the RNA-Polymerase as a damage sensor, transcription and DNA repair are directly coupled, and so the whole process is commonly referred to as ‘transcription coupled repair’ (TCR). Approaching a DNA damage site leads to a stopped RNAP, a so-called ‘stalled transcription complex’, at these sites. Mfd (mutation frequency decline) was initially identified as a key TCR-factor in Escherichia coli (E. coli) over 20 years ago (Selby and Sancar, 1993), and this discovery let to Aziz Sancar winning the Nobel Prize for Chemistry in 2015 together with two other pioneers in the field of DNA repair: Thomas Lindahl and Paul Modrich. It could be shown that Mfd uses its ATPase function to move along the DNA to displace RNAP from the DNA template.
Next, Mfd is able to recruit the DNA-repair machinery to the DNA-lesion to initiate its repair. Nevertheless, the mild sensitivity to UV-radiation and DNA-damaging agents to bacteria lacking Mfd raised doubts early on about whether this pathway might be the sole responsible cellular process.
Beyond Mfd
The latest studies have shown that two other important proteins regulating transcription and DNA repair, NusA and UvrD, might be alternative recruitment factors as they contribute significantly to TCR via Mfd-independent pathways (Belogurov and Artsimovitch, 2015). The importance of these newly discovered pathways is evidenced on the genetic level as nusA and uvrD mutants show high sensitivity to UV-induced DNA-damage. While the functional details of these TCR pathways remain to be discerned, available data indicates that their function relies on a completely different mechanism than Mfd as they either stabilise backtracked states or actively pull RNAP backwards and away from the DNA-lesion to facilitate DNA-repair and subsequently initiate a restart of transcription.
In our research we are now focusing on the structural and functional details of the involvement of UvrD and NusA in these repair pathways with the aim of elucidating the detailed roles of all three proteins (MfD, UvrD, and NusA) in order to discern the basis of their overlapping functionalities.
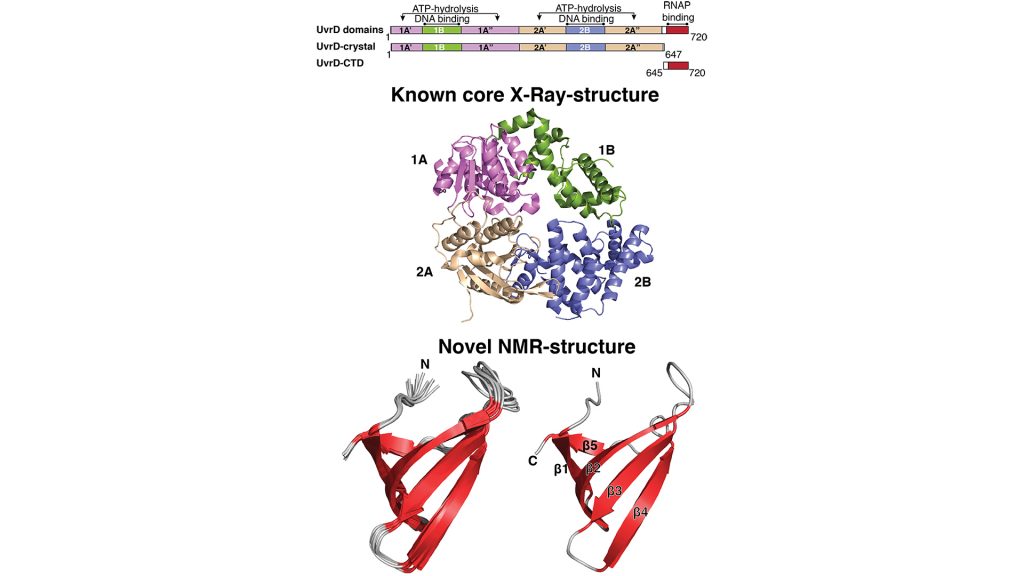
Recently, our research on the structure and function relationship of the UvrD-helicase delivered important new insights. While the crystal structure of UvrD has already been known for some time, there was an outstanding question: are the last ~90 residues of the protein structured in some way, and how might they contribute to the functionality of the protein?

Tudor domains
Because crystallographic studies indicated that these residues were largely unstructured, they had previously been omitted from structural investigations. By using solution NMR-spectroscopy, my team was able to not only show that this region is not unstructured, but also that it folds into a small so-called ‘β-barrel’ structure, called a Tudor domain (Kawale and Burmann, 2020). These Tudor domains are very characteristic for protein and nucleic acid, e.g. DNA or RNA, binding domains that are found in all kingdoms involved in a variety of nucleic acid based processes, like replication and transcription.
Remarkably, our studies revealed that this domain shows high plasticity in its properties to bind to very different binding partners, such as the RNA Polymerase as well as DNA, which it achieves by sophisticated structural modulations. Our work indicates that the ambiguities in its interaction partners might be the key to the role of this domain as a binding hub, which might also be crucial for the recruitment of DNA-repair machinery, a feature my team is currently investigating. Therefore, we hope that our research will contribute significantly to the detailed understanding of this essential DNA-repair pathway, filling the still existing blanks in our understanding of these fundamental process.
References
Burmann, B. M. et al. ’A NusE:NusG complex links transcription and translation’. Science 328, 501–504 (2010)
Truglio, J. J., Croteau, D. L., Van Houten, B. and Kisker, C. ‘Prokaryotic nucleotide excision repair: the UvrABC system’. Chem. Rev. 106, 233–252 (2006)
Sancar, A. and Rupp, W. D. ‘A novel repair enzyme: UvrABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region’. Cell 33, 249–260 (1983)
Selby, C. P. and Sancar, A. ‘Molecular mechanism of transcription-repair coupling’. Science 260, 53–58 (1993)
Belogurov, G. A. and Artsimovitch, I. ‘Regulation of Transcript Elongation’. Annu. Rev. Microbiol. 69, 49-69 (2015)
Kawale, A. A. and Burmann, B. M. ‘UvrD helicase-RNA polymerase interactions are governed by UvrD’s carboxy-terminal Tudor domain’. Commun Biol 3, 607 (2020)
Acknowledgements
Continuous funding from the Swedish Research Council (Vetenskapsrådet) and the Knut och Alice Wallenberg Foundation through a Wallenberg Academy Fellowship, as well as through the Wallenberg Centre for Molecular and Translational Medicine, University of Gothenburg, Sweden, is gratefully acknowledged. The Swedish NMR Centre of the University of Gothenburg is acknowledged for spectrometer time.

Please note, this article will also appear in the fifth edition of our quarterly publication.