Dr Johannes Thoma and Dr Björn Burmann discuss their work using outer membrane vesicles (OMVs) to potentially gain new insight into antibiotic resistance mechanisms.
One of the most acute threats to global health is the rapid spread of antimicrobial resistance combined with a lack of new antibiotic medication. Among multi-resistant bacteria, Gram-negative bacteria such as diverse species of Salmonella, Pseudomonas, and Acinetobacter are particularly challenging. The reason is that Gram-negative bacteria are surrounded by a complex cell envelope, which consists of two lipid membranes – an inner and an outer membrane. The space between the two membranes, called the ‘periplasm’, contains the bacterial cell wall, which is formed by a stable network of amino acids and sugar molecules, called ‘peptidoglycan’ (Silhavy, Kahne, and Walker, 2010).
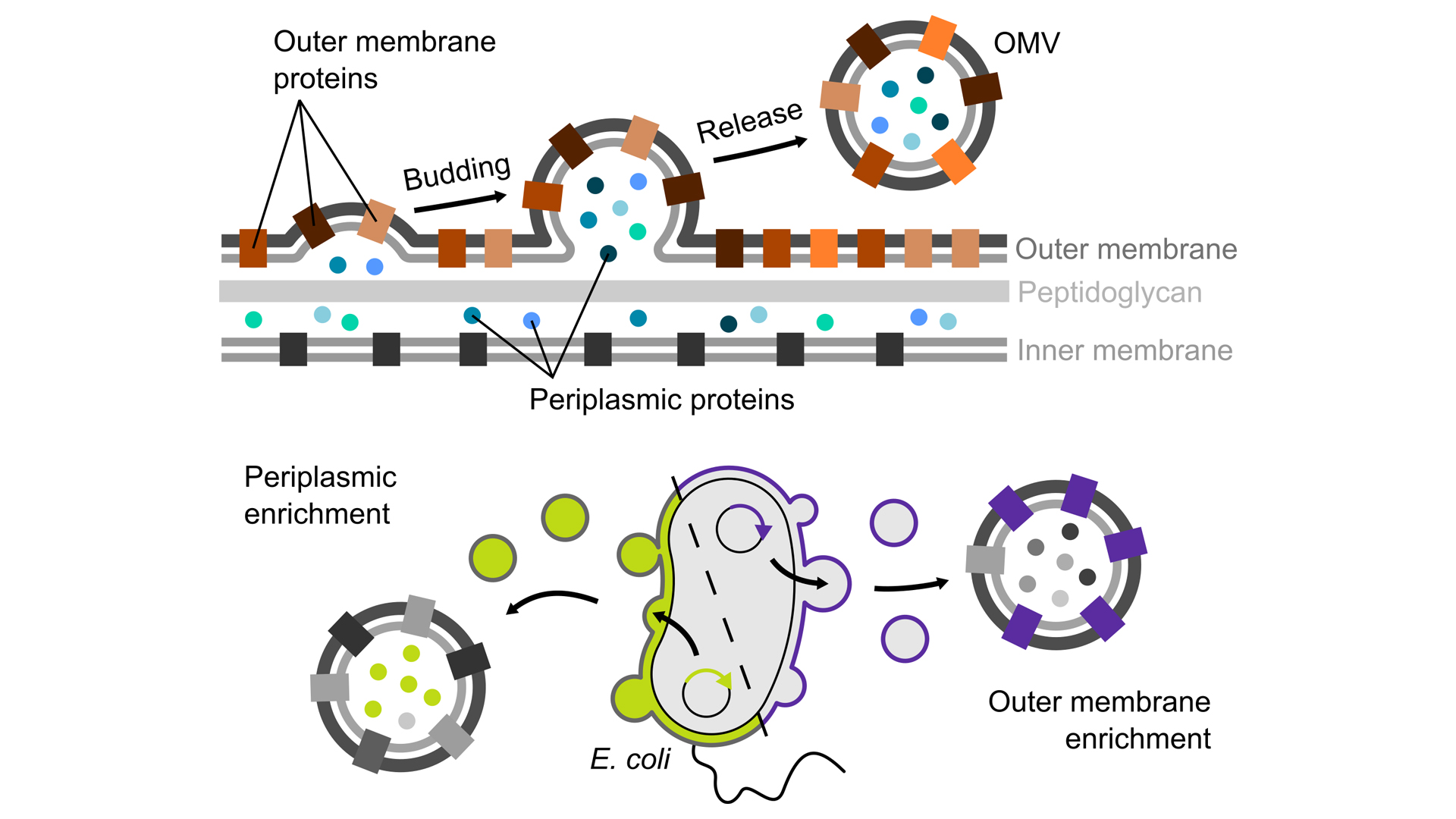
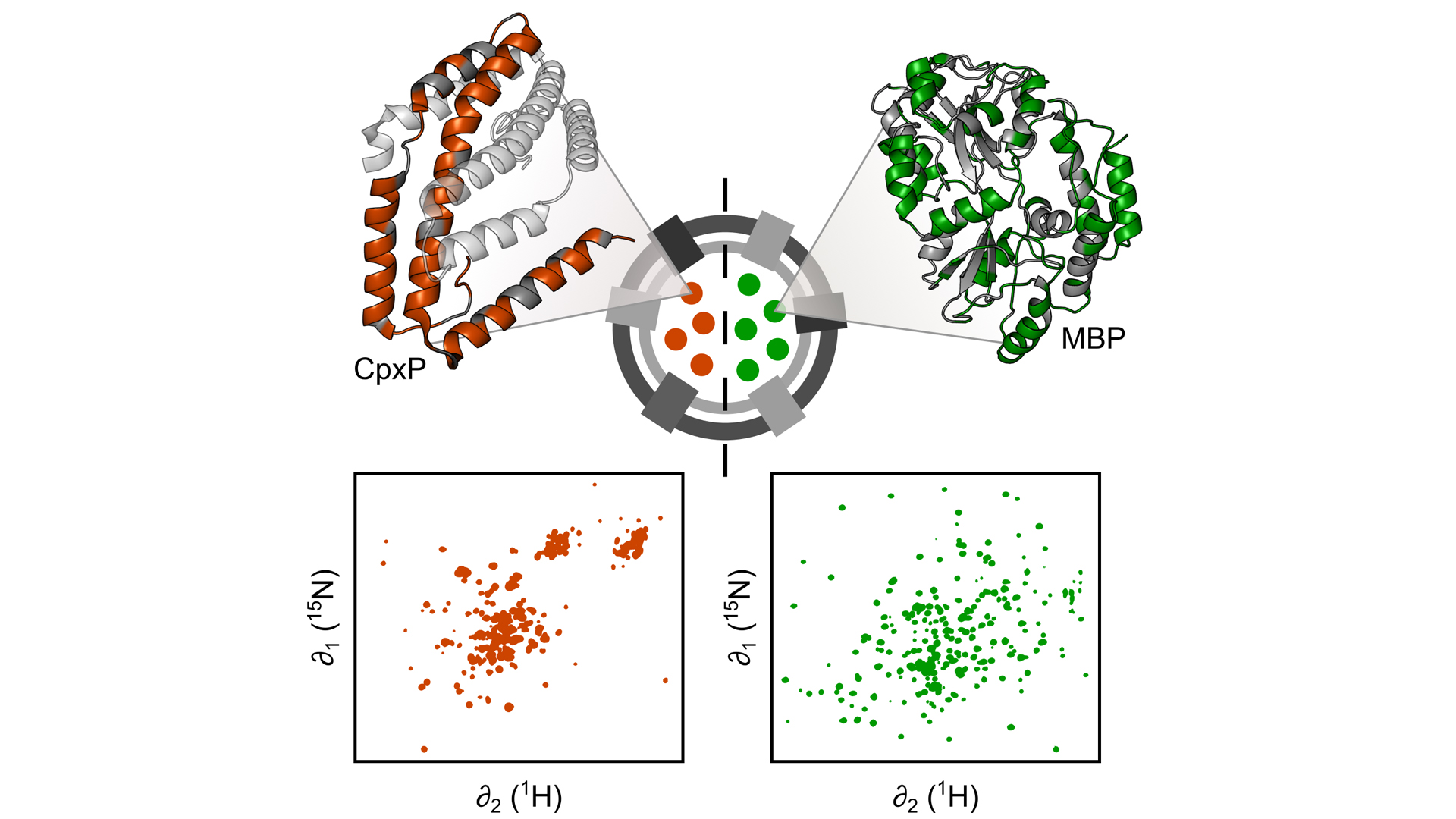
Fig. 2: Engineered OMVs are excellent tools for studying periplasmic proteins, for example CpxP (left), a stress-sensing protein, and the maltose binding protein (MBP, right), under ‘real-life’ conditions without interference from the remaining, much larger remaining bacterial cell. To this end, we use nuclear magnetic resonance (NMR) spectroscopy, which allows us to characterise the structure and dynamics of periplasmic proteins encapsulated in OMVs at atomic resolution
While the bacterial inner membrane is a common lipid membrane, similar to the cell membranes of all living organisms, the outer membrane forms an additional protective barrier and is a unique property of gram-negative bacteria. Bacterial outer membranes contain specialised large molecules, so-called ‘lipopolysaccharides’ (LPS), which consist of a lipid nucleus and a long carbohydrate chain built up from a diversity of sugar molecules. A stable layer of LPS makes the outer membranes of gram-negative bacteria almost impermeable to a majority of molecules, including many antibiotics.
The outer membrane as a barrier
The transport of molecules across the outer membrane barrier is regulated by pore-forming membrane proteins. These proteins span the membrane in the form of open barrels (hence their name ‘transmembrane β-barrels’) and allow the passage of only small molecules. Therefore, only small, water-soluble antibiotic medications such as penicillin and its derivatives, a class called ‘β-lactam antibiotics’, are effective against Gram-negative bacteria. Larger molecules cannot cross the outer membrane barrier and thus never reach the inside of the bacterial cell. The outer membrane thus makes Gram-negative bacteria naturally resistant to most large molecular antibiotics (Nikaido, 2003). β-lactam antibiotic medicine’s work by interfering with the synthesis of the bacterial cell wall embedded in the periplasm. As a second line of defense against antibiotics, resistant bacteria have specialised enzymes residing in their periplasm, so-called ‘β-lactamases’, which inactivate β-lactam antibiotics (Tooke et al., 2019).
An isolated cell envelope microcosm
The complex architecture of the bacterial cell envelope makes it not only complicated for antibiotic treatments to reach the cell interior, but also to study its functional details with biophysical methods, especially since the cell envelopes make up only 10% of the cellular volume. Until now, it has been impossible to recreate this particular cellular environment in the laboratory to characterise it in detail. Thus, the majority of current knowledge about envelope components has been obtained based on purified components, which have been isolated from bacterial cells. However, almost nothing is known about how envelope components, such as proteins, behave in and together with their natural cellular environment – a large gap in knowledge, the closing of which will help to better tackle antimicrobial resistance.
Fortunately, in a natural process, Gram-negative bacteria bud off parts of their cell envelopes in the form of small blisters, called ‘outer membrane vesicles’ (OMVs). These vesicles are surrounded by the bacterial outer membrane and are filled with periplasmic components. The vesicles can therefore be seen as an isolated microcosm, exclusively consisting of intact cell envelope components.
Engineered OMVs as research tools
In our on-going research, we develop methods that make it possible to use OMVs as nanotools in the lab by manipulating the composition of these vesicles towards our research questions. To this end we use specialised Escherichia coli strains, which produce large amounts of OMVs and which allow us to specifically target the proteins we are interested in to these vesicles (Thoma, Manioglu, et al., 2018). Such engineered OMVs are excellent tools for studying biological phenomena involving the bacterial cell envelope, without interference from the remaining, much larger bacterial cell. Using OMVs for the first time allows us to characterise how proteins in the cell envelope interact with each other and how their structure and function are affected by the other components of the cell envelope. For example, the native membrane of OMVs allowed us to characterise how the unique lipid composition of the bacterial outer membrane and its LPS molecules influence the stability of proteins embedded in this membrane (Thoma, Sun, et al. 2018).
Molecular details under ‘real-life’ conditions
More recently, we used OMVs to characterise the structure and dynamics of periplasmic proteins encapsulated in OMVs using nuclear magnetic resonance (NMR) spectroscopy at atomic resolution (Thoma and Burmann, 2020). Our work shows that, despite subtle differences, for many proteins the overall 3D structure is similar to what has previously been determined with purified proteins. However, some proteins appear to have a vastly different structure in their natural environment, unlike anything observed with purified proteins before. Moreover, our work indicates that the dynamic behaviour of proteins is directly influenced by the dense periplasmic environment, which potentially impacts how they catalyse chemical reactions. Thus, with our new methodology we now have a unique toolset at hand, which allows us to understand the molecular details of how periplasmic processes occur under ‘real-life’ conditions. Using OMVs, we now hope to gain new insight into antibiotic resistance mechanisms, e.g. how β-lactamases behave in the natural periplasm – knowledge which is the key to developing new antibiotic medications against Gram-negative bacteria.
Acknowledgment
Continuous funding from the Swedish Research Council (Vetenskapsrådet) and the Knut och Alice Wallenberg Foundation through a Wallenberg Academy Fellowship, as well as through the Wallenberg Centre for Molecular and Translational Medicine, University of Gothenburg, Sweden is gratefully acknowledged. The Swedish NMR Centre of the University of Gothenburg is acknowledged for spectrometer time.



References
- Hiroshi Nikaido. 2003. ‘Molecular Basis of Bacterial Outer Membrane Permeability Revisited’. Microbiology and Molecular Biology Reviews: MMBR 67 (4): 593–656. https://doi.org/10.1128/MMBR.67.4.593
- Thomas J. Silhavy, Daniel Kahne, and Suzanne Walker. 2010. ‘The Bacterial Cell Envelope’. Cold Spring Harbor Perspectives in Biology 2 (5): 1–16. https://doi.org/10.1101/cshperspect.a000414
- Thoma, Johannes, and Björn M. Burmann. 2020. ‘High-Resolution In Situ NMR Spectroscopy of Bacterial Envelope Proteins in Outer Membrane Vesicles’. Biochemistry 59 (17): 1656–60. https://doi.org/10.1021/acs.biochem.9b01123
- Johannes Thoma, Selen Manioglu, David Kalbermatter, Patrick D. Bosshart, Dimitrios Fotiadis, and Daniel J. Müller. 2018. ‘Protein-Enriched Outer Membrane Vesicles as a Native Platform for Outer Membrane Protein Studies’. Communications Biology 1 (1): 23. https://doi.org/10.1038/s42003-018-0027-5
- Johannes Thoma, Yang Sun, Noah Ritzmann, and Daniel J. Müller. 2018. ‘POTRA Domains, Extracellular Lid, and Membrane Composition Modulate the Conformational Stability of the ß-Barrel Assembly Factor BamA’. Structure 26 (7): 987-996.e3. https://doi.org/10.1016/j.str.2018.04.017
- Catherine L. Tooke, Philip Hinchliffe, Eilis C. Bragginton, Charlotte K. Colenso, Viivi H.A. Hirvonen, Yuiko Takebayashi, and James Spencer. 2019. ‘ß-Lactamases and ß-Lactamase Inhibitors in the 21st Century’, Journal of Molecular Biology 431 (18): 3472–3500. https://doi.org/10.1016/j.jmb.2019.04.002
Please note, this article will also appear in the sixth edition of our quarterly publication.









